 Back Back
Research Article
ScienceAsia 30 (2004): 239-245 |doi: 10.2306/scienceasia1513-1874.2004.30.239
Selection of Y-Chromosomal Microsatellites for
Phylogenetic Study Among Hilltribes in Northern
Thailand Using the Decision Tree Induction Algorithm
Daoroong Kangwanpong,a,* Jeerayut Chaijaruwanich,b Metawee Srikummoola and Jatupol Kampuansaia
ABSTRACT: A computational science approach, based on genetic information and methods, was utilized to
calculate the minimum number and select the most suitable microsatellite markers for evaluating the
diversity and genetic distance among the hilltribes in Northern Thailand. Selecting from previously available
and newly published microsatellites, we studied genetic variation and distance - Fst and (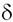 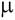 )2, of 4 hilltribe
populations in Northern Thailand using 15 universal known loci of Y-chromosomal microsatellites. The
Decision Tree Induction algorithm in information theory was used to measure impurity of categorizing
populations by the number of tandem repeats at each loci and select the minimum number of microsatellites
markers with the most discriminating power. Seven selected microsatellites, 8 unselected microsatellites, the
original 15 markers, and another four haplotypes used in different global population studies were then
employed to construct the UPGMA trees. To validate our results, the Relative Optimality Score (R value)
was calculated from the total branch length of the trees, using the tree constructed from genetic distances
obtained from 15 Y-chromosomal microsatellites as reference. The graph between R values and different
haplotypes was plotted and compared between Fst and ( )2, of 4 hilltribe
populations in Northern Thailand using 15 universal known loci of Y-chromosomal microsatellites. The
Decision Tree Induction algorithm in information theory was used to measure impurity of categorizing
populations by the number of tandem repeats at each loci and select the minimum number of microsatellites
markers with the most discriminating power. Seven selected microsatellites, 8 unselected microsatellites, the
original 15 markers, and another four haplotypes used in different global population studies were then
employed to construct the UPGMA trees. To validate our results, the Relative Optimality Score (R value)
was calculated from the total branch length of the trees, using the tree constructed from genetic distances
obtained from 15 Y-chromosomal microsatellites as reference. The graph between R values and different
haplotypes was plotted and compared between Fst and (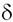 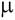 )2 genetic distances. The results show that the
haplotype of 7 selected markers is the most reliable compared to all other haplotypes, while the other 8
nonselected loci may be unnecessary for determining the genetic distance in our study. Using the decision
tree induction algorithm we were able to select the microsatellites with three levels of discriminating power
for differentiating populations, and thereby could reduce the number of Y-chromosomal microsatellite used
to half and still achieve the same information. )2 genetic distances. The results show that the
haplotype of 7 selected markers is the most reliable compared to all other haplotypes, while the other 8
nonselected loci may be unnecessary for determining the genetic distance in our study. Using the decision
tree induction algorithm we were able to select the microsatellites with three levels of discriminating power
for differentiating populations, and thereby could reduce the number of Y-chromosomal microsatellite used
to half and still achieve the same information.
Download PDF
a Genetics and Molecular Biology Unit, Department of Biology, Faculty of Science,
Chiang Mai University, Chiang Mai 50202, Thailand.
b Department of Computer Science, Chiang Mai University, Chiang Mai 50202, Thailand.
* Corresponding author, E-mail: scidkngw@chiangmai.ac.th
Received 9 Jan 2004,
Accepted 13 Jul 2004
|

